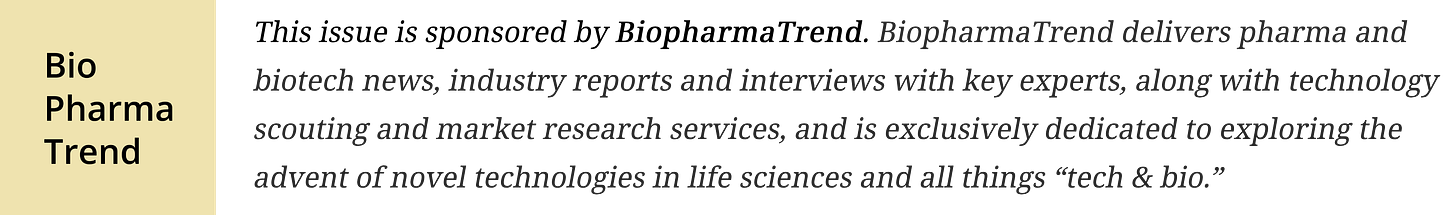Weekly Tech+Bio Highlights #55: AI-Generated Functional Bacteriophages
Over $100M in funding & early clinical updates on AI drugs
Hi! This is BiopharmaTrend’s weekly newsletter, Where Tech Meets Bio, where we explore technologies, breakthroughs, and cutting-edge companies.
If this newsletter is in your inbox, it’s because you subscribed, or someone thought you might enjoy it. In either case, you can subscribe directly by clicking this button:
🤖 AI x Bio
(AI applications in drug discovery, biotech, and healthcare)
🔹 Stanford and Arc Institute researchers used an AI genome model to design 302 synthetic bacteriophage genomes ΦX174, with 16 successfully replicating in E. coli, demonstrating the potential of generative AI for engineering therapeutic functional viruses.
🔹 NVIDIA supports the UK’s sovereign AI strategy through wide-ranging collaborations across academia and biotech, advancing AI models like Nightingale for early diagnosis, Basecamp’s massive bio-foundation datasets, Peptone’s physics-driven protein targeting, and Isomorphic Labs’ AlphaFold-based drug design engine, and many others.
🔹 Revvity partners with Profluent to integrate AI-designed adenine deaminases into its CRISPR-based Pin-point base editing platform, enhancing precision, flexibility, and licensing access for therapeutic gene editing applications.
🔹 Enveda begins Phase 1b trial of ENV-294, an AI-discovered, first-in-class small molecule for atopic dermatitis, following strong safety data in healthy volunteers and leveraging its PRISM platform for accelerated drug development.
🔹 MEDITECH previews new AI initiatives at MEDITECH LIVE 2025, including agentic user experiences within its Expanse EHR, featuring clinician and patient chatbots, voice interactions, and generative AI tools aimed at improving care delivery.
🔹 Switzerland launches NAIPO, an Innosuisse Flagship Initiative co-led by EPFL and ETH, backed by CHF 18.9M to develop national AI infrastructure for precision oncology, uniting 21 partners to accelerate cancer diagnosis, treatment, and research translation.
🔹 Sage Bionetworks and Biomni partner to enable AI-powered natural language search across 3 petabytes of biomedical data via Synapse, streamlining access to fragmented datasets in oncology, neuroscience, and rare diseases while maintaining strict data governance.
🔹 Sapio Sciences launches ELaiN, allegedly the world’s first AI-powered 3rd-generation Electronic Lab Notebook, introducing an agentic co-scientist that helps design, execute, and analyze experiments across R&D domains like bioinformatics and cheminformatics.
🔹 Rubedo Life Sciences receives FDA clearance to begin a Phase 1b/2a trial of RLS-1496, a first-in-class GPX4 modulator targeting senescent cells in actinic keratosis, expanding its AI-driven anti-aging pipeline already in trials for psoriasis and atopic dermatitis.
🔹 ProteinQure doses first patient in Phase I trial of PQ203, an AI-designed peptide therapeutic targeting metastatic solid tumors, marking a milestone for computational drug discovery and advancing treatment options for resistant triple-negative breast cancer.
🔹 Formation Bio expands its leadership team with top biotech and pharma executives from Pfizer, BenevolentAI, and Incyte to scale its AI-native drug development model, aiming to accelerate asset acquisition, clinical advancement, and out-licensing growth.
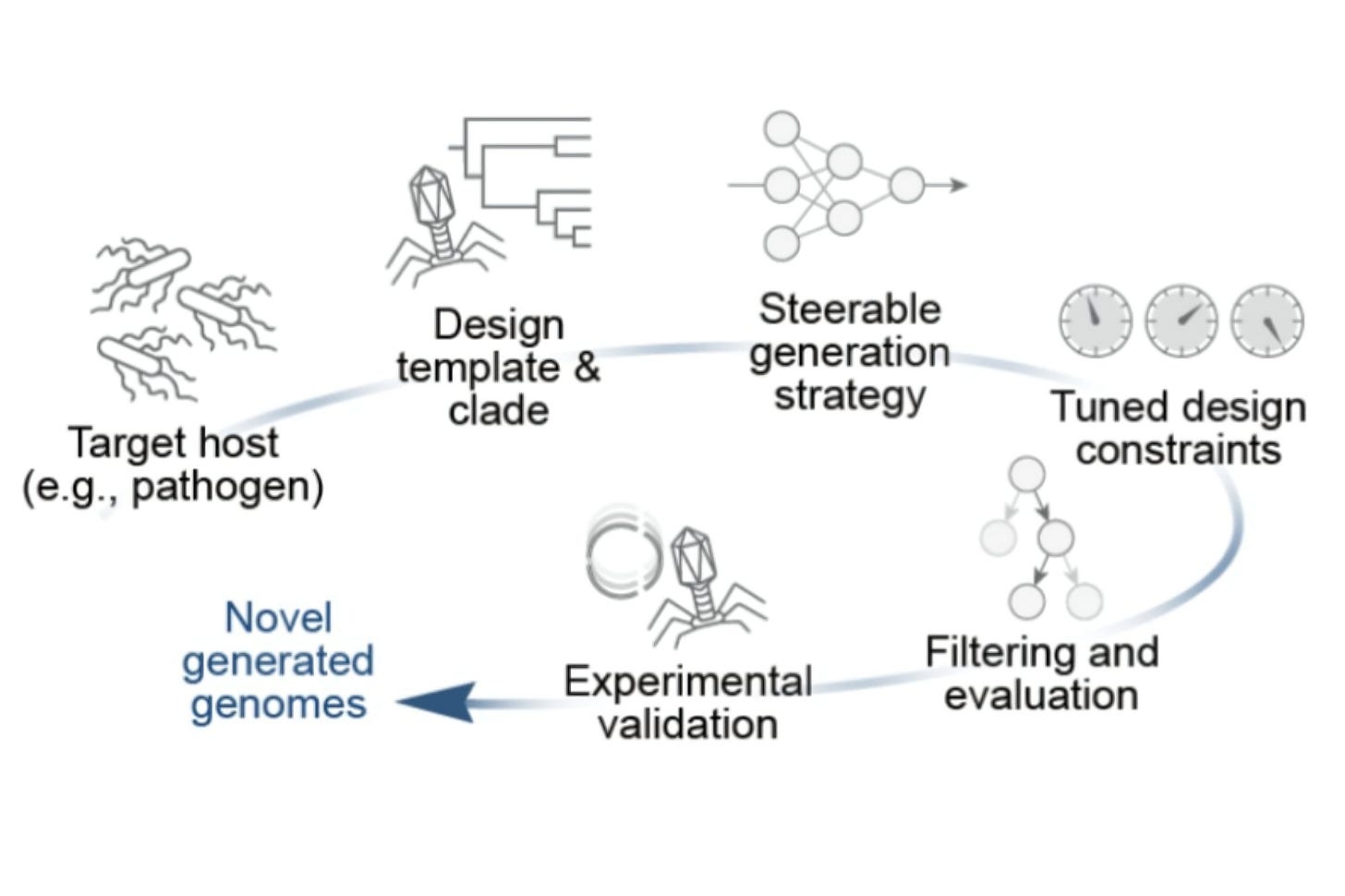
🔹 Prescient Design and Genentech introduce IGLOO, a multimodal tokenizer for antibody loop structures that combines sequence and backbone dihedral angle information, achieving state-of-the-art accuracy in CDR clustering, binding affinity prediction, and structure-guided antibody loop generation.
🔹 Insilico Medicine partners with Capgemini to integrate AI-driven drug discovery with advanced digital infrastructure, focusing on foundational AI models, automated labs, and rapid discovery sprints to streamline early-stage R&D.
🚜 Market Movers
(News from established pharma and tech giants)
🔹 Bayer advances two potential disease-modifying therapies for Parkinson’s, launching a Phase III trial for its stem cell therapy bemdaneprocel and a Phase II trial for gene therapy AB-1005, marking the first company to push both approaches in parallel into late-stage development.
🔹 Roche acquires 89bio for up to $3.5B, gaining late-stage FGF21 analog pegozafermin for MASH, as the Swiss pharma aims to compete with Madrigal and Novo Nordisk in the fast-growing metabolic liver disease market.
🔹 Infinitus teams up with IBM Consulting to deploy AI agents for automating benefit verification and prior authorization in specialty pharmacy, aiming to reduce administrative delays and accelerate patient access to therapies.
💰 Money Flows
(Funding rounds, IPOs, and M&A for startups and smaller companies)
🔹 T.Rx Capital raises $77.5M for its first fund to invest in early-stage biotech and digital health companies, with a focus on techbio ventures and support from high-profile industry leaders including Robert Langer.
🔹 Immuto raises $8M in seed funding to advance its AI-driven structural surfaceomics platform for cell-surface target discovery, and enters a drug discovery collaboration with Daiichi Sankyo focused on novel antibody targets in solid tumors.
🔹 Trially AI raises $4.7M in seed funding led by Flyover Capital and launches Margo, an AI-powered platform aimed at accelerating clinical trial enrollment by automating patient matching, engagement, and pre-screening.
🔹 Innovaccer acquires Story Health to expand its AI capabilities in specialty care, introducing agentic AI systems that support continuous patient engagement and assist care teams in reducing hospitalizations and improving treatment adherence.
🔹 Salt AI raises $10M led by Morpheus Ventures to scale its drag-and-drop AI workflow platform for life sciences, enabling biopharma and healthcare teams to rapidly build, version, and deploy multi-model pipelines for drug discovery and clinical operations.
⚙️ Other Tech
(Innovations across quantum computing, BCIs, gene editing, and more)
🔹 University of Bath spinout Prothea Technologies launches a clinical trial in Edinburgh for a novel optical fibre endoscope designed to improve lung cancer biopsies, enabling real-time imaging and potentially same-visit diagnosis and treatment.
This newsletter reaches over 9.6K industry professionals from leading organizations across the globe. Reserve your sponsor slot in one of the upcoming issues.
Contact us at info@biopharmatrend.com
Generating Viral Genomes
Arc Institute and Stanford researchers have taken a step toward what they call “genome design,” using large language models to generate synthetic viral genomes that actually work in the lab. In a preprint posted last week, the team reports creating 16 viable bacteriophages (viruses that infect bacteria) generated by fine-tuned versions of their Evo 1 and Evo 2 genome models (we covered Evo 2 when it came out in February this year). The base Evo models were trained on 2.7 million raw prokaryotic and phage genome sequences, and Evo 2’s corpus spans 9.3 trillion nucleotides from 128,000 organisms including bacteria, archaea, and eukaryotes. Current work focuses on ΦX174, a historically significant phage that’s compact but packed with complexity due to overlapping genes and regulatory regions.
As for the novelty, Evo has already been used to generate proteins and multi-component systems like CRISPR complexes. But full genomes introduce failure modes that aren’t an issue with isolated proteins, especially when dealing with architectures like ΦX174, where one mutation can knock out multiple genes at once.
To make this viable, Arc developed a full stack: a custom gene annotation pipeline that could handle overlapping reading frames, fine-tuning of Evo on ~14,000 Microviridae sequences to bias outputs toward ΦX174-like designs, and a high-throughput validation workflow using growth inhibition assays in E. coli.
From 302 model-generated genomes, they synthesized and tested 285. 16 successfully booted up in E. coli C and W (standard, non-pathogenic lab strains), with one—Evo-Φ2147—diverging enough from known sequences to potentially qualify as a new species. In some cases, like Evo-Φ36, the AI incorporated protein swaps from distantly related phages that had previously failed; here it worked, and electron microscopy shows the shorter protein sits differently in the capsid, with other sequence changes enabling the combo.
Researchers also tested whether this design diversity could help overcome bacterial resistance. In E. coli strains engineered to resist ΦX174, the team’s phage cocktails (consisting of combinations of AI-generated variants) restored infectivity within a few passages. Wild-type ΦX174 failed completely.
The results point to an advantage in using generative models to explore genome-scale sequence space at volume. Instead of waiting for nature to evolve phages that match specific constraints, the team generated hundreds of diverse candidates and screened for the ones that worked, with some being functionally superior to wild-type (case in point, Evo-Φ69 rose ~65x in mixed competitions).
While the biology of the phages didn’t fundamentally change and they still infected the same hosts, the ability to reliably generate viable variants with significant novelty may signal a shift in how synthetic virology is done. As Niko McCarty put it in his writeup for Asimov Press, this may be “more of a technical milestone than a practical one,” but it sets up future work where genome-scale design could target traits beyond viability—like tropism, expression dynamics, or immune evasion:
“It’s also worth asking whether this kind of full-genome design is as useful for bioengineering as it initially seems. In many cases, especially with viruses, a handful of genes drive the traits that matter most. Adeno-associated viruses, for example, infect specific tissues in the human body due to particular proteins embedded in their capsids. By tinkering with the genes encoding those proteins, one can alter which cells these viruses infect. And so, with this logic in mind, perhaps the wholesale, bottom-up design of an entire phage genome may be more of a technical milestone than a practical one. Using AI to design single genes or regulatory regions — where you know which knobs you’re turning — may, in many cases, be the more efficient strategy.”
Here’s a security caveat—the Evo 2 model is open-source (code, weights, data), and while human viruses were excluded from training, the underlying method is adaptable. Safety language excludes pathogens that infect humans and other complex organisms and configures the model not to return productive answers on those. For now, the barrier isn’t the model itself but rather the data and the cost of synthesis at larger scales. According to Asimov Press, DNA synthesis runs about $0.07/base at Twist Biosciences and can reach ~$0.45/base for difficult builds; Cambridge’s recoded 57-codon E. coli reportedly took several years and cost >$1M.
Tokenizing Antibody Loops
Antibody binding is largely determined by the complementarity-determining regions (CDRs), which form flexible loop structures. Antibody “loops” are short, flexible segments at the tips of the variable regions that actually touch the antigen; they largely set binding specificity, with H3 (heavy-chain CDR3) usually the most diverse in length and shape. These loops are key to understanding antigen interactions and development of novel drugs. Because these loops do the binding work, representing them cleanly helps models compare similar shapes, predict affinity changes, and propose loop designs that keep the right geometry.
A recent preprint from Fang, Alberstein, Kelow, and Dreyer introduces IGLOO (ImmunoGlobulin LOOp Tokenizer), a multimodal tokenizer that represents antibody CDR loops encoding both sequence and backbone dihedral angles. It’s trained with a contrastive objective so loops with similar backbones land close together in latent space. The reported benefits: better retrieval of structurally similar loops (notably H3 - antibody loop type), full coverage beyond canonical clusters, and practical drop-ins for language models that help with affinity prediction and loop sampling. The model architecture can be seen below:
The model learns embeddings where structurally similar loops sit close together, and it produces both continuous and discrete tokens.
What stood out:
Coverage: IGLOO assigns tokens to every loop, while still recovering ~90% of known canonical conformations.
Retrieval: It retrieves closer structural matches than previous methods, improving H3 loop retrieval by about 6%.
Language models: Adding IGLOO tokens to antibody LMs boosted binding affinity prediction (better performance on 8/10 benchmarks) and supported loop sampling that stayed structurally consistent even with diverse sequences.
The authors note that results are so far in silico, and more wet-lab validation will be needed. Still, IGLOO shows how moving from residue-level to loop-level tokens can make antibody language models more structure-aware and potentially more useful for design tasks.