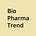Weekly Tech+Bio Highlights #43: New Foundation Model from MIT & Recursion
Also: Spatial Biology Means Terabyte-Scale Files, A Visual Guide to Genome Editors, Biotech VC Enters Quieter Phase, & Why AI Hallucinations May Be a Topological Problem
This week saw the FDA launch Elsa for internal drug review, MIT and Recursion release an open-source model for both protein structure and affinity prediction, and FutureHouse report AI-driven discovery of a dry AMD candidate. Insilico Medicine shared Phase IIa results for an AI-designed fibrosis drug in Nature Medicine, while ImmunoPrecise mapped a pan-dengue vaccine target fully in silico.
The London Biotechnology Show returns June 18-19, featuring themes in AI-driven bioprocessing, cell and gene therapy, and digital infrastructure—stay tuned for our post-event coverage!
The highlights above are a quick summary—hyperlinked stories with more detail are organized in the sections below.
Hi! This is BiopharmaTrend’s weekly newsletter, Where Tech Meets Bio, where we explore technologies, breakthroughs, and cutting-edge companies.
If this newsletter is in your inbox, it’s because you subscribed, or someone thought you might enjoy it. In either case, you can subscribe directly by clicking this button:
🤖 AI x Bio
(AI applications in drug discovery, biotech, and healthcare)
🔹 The FDA launched Elsa, a generative AI platform to assist staff across drug review, inspection, and safety workflows to accelerate clinical protocol reviews, label comparisons, and adverse event summarization.
🔹 MIT’s Jameel Clinic and CSAIL, with Recursion, release Boltz-2, an open-source biomolecular model that jointly predicts protein structure and binding affinity, on par with FEP accuracy while running 1000x faster, as per company claim.
🔹 AI-designed lung fibrosis therapy shows early signals—Nature Medicine publishes Phase IIa results for Insilico’s AI-developed TNIK inhibitor, with a high-dose group showing a +98.4 mL lung function gain vs decline for placebo.
🔹 Harvard researchers publish a foundation model for multiplex spatial proteomics, trained on 47 million image patches spanning 175 protein markers, 16 tissue types and 8 imaging platforms.
🔹 AI maps universal vaccine target for dengue—ImmunoPrecise used in silico methods to find a shared weak spot across all four dengue virus types, potentially enabling a broad vaccine.
🔹 Roadmap to fight superbugs—César de la Fuente and colleagues publish a tutorial in Nature Protocols outlining a machine learning playbook for mining genomes and proteomes to discover new antibiotics, aiming to accelerate solutions to antimicrobial resistance entirely from biological data.
🔹 Google DeepMind and the Fleming Initiative launch a three-year academic fellowship at Imperial College London to support early-career researchers applying AI to antimicrobial resistance; applications are open until July 1st.
🔹 FutureHouse reports that its multi-agent AI system autonomously generated and iterated on hypotheses, ultimately flagging the approved ripasudil as a novel candidate therapy for dry age-related macular degeneration. They’ve also released ether0, a 24B open-weight reasoning model for molecular design.
🔹 Meta’s FAIR team and collaborators release Open Molecules 2025 (OMol25), a dataset of 100M+ DFT calculations across 83M molecules, enabling high-accuracy ML models for energy prediction in biomolecules, catalysts, electrolytes, and metal complexes.
🔹 In a recent blog post, mathematician and microbiology PhD student Rachel Thomas (fast.ai co-founder) contrasts two enzyme function studies to spotlight how flashy deep learning results (despite serious biological errors) are celebrated, while meticulous fact-checking by domain experts goes largely ignored; calls for stronger integration of domain expertise in machine learning research.
🔹 Medidata and Medable unveil new digital trial tools at ASCO—Medidata launches an AI-powered protocol simulator to improve study design, while Medable debuts a virtual follow-up model to streamline long-term tracking in cell and gene therapy trials.
🔹 Reflections on building TechBio—Recursion CEO Chris Gibson joins the Good Medicine podcast hosted by David Bearss Ph.D to discuss the company’s 11-year journey scaling AI-driven drug discovery, emphasizing the long-term vision of mapping biology, shifting toward in silico development, and reshaping medicine to prevent rather than just treat disease.
🔹 Chinese AI firm DeepSeek begins hiring interns with medical backgrounds to label data and reduce hallucinations in its diagnostic models, as its LLMs see growing use in over 300 hospitals across China despite safety concerns from researchers.
🔹 The FDA has granted de novo clearance to Clairity for the first AI tool that predicts five-year breast cancer risk from screening mammograms, validated on over 77,000 cases.
🚜 Market Movers
(News from established pharma and tech giants)
🔹 Sarepta Therapeutics (est. in 1980) received first FDA Platform Technology Designation for its rAAVrh74 viral vector.
💰 Money Flows
(Funding rounds, IPOs, and M&A for startups and smaller companies)
🔹 AI-biotech deals now routinely fetch $20-65M upfront, milestones top $1B—in Nature Biopharma Dealmakers Andrew Marshall reports that AI-focused biopharma partnerships continue to grow in scale and scope. Companies expand beyond small-molecule discovery into biologics, oligonucleotides, and data licensing.
🔹 Scaling drug ingredients with biosynthesis—Antheia raises $56M Series C (led by GHIC and EDBI) to commercialize biosynthetic production of key drug ingredients like thebaine, expand into Asia, and boost supply chain resilience amid global drug shortages.
🔹 Amplify Partners launches a $200M biotech-focused fund to back early-stage digital biology startups blending lab science with AI and computation, with plans to make ~20 investments.
🔹 UK government announces £86B R&D investment through 2029, including £500M for regional innovation, to accelerate progress in AI, biotech, clean energy, and advanced manufacturing, aiming to drive economic renewal and create high-skilled jobs across the country.
⚙️ Other Tech
(Innovations across quantum computing, BCIs, gene editing, and more)
🔹 Huang et al. engineered the auditory protein prestin to mimic echolocating species, enabling ultrasound-triggered calcium signaling and wireless neurostimulation in deep brain regions, offering a noninvasive tool for modulating cellular activity.
🔹 Wyss Institute scientists create a new protein-based nanoparticle drug delivery system that helps medicines escape cellular traps, allowing gene editors like CRISPR to reach their target more effectively—achieving up to 83% editing efficiency in mice with low toxicity.
🔹 MIT grad student Yehlin Cho releases BoltzDesign1, a protein design tool that inverts Boltz-1 (“an open-source reproduction of AlphaFold3”) to generate atomic-level binders for molecular targets without model finetuning.
🏛️ Bioeconomy & Society
(News on centers, regulatory updates, and broader biotech ecosystem developments)
🔹 Cancer research at risk—ASCO CEO Clifford Hudis warns that proposed 40% cuts to NIH and NCI funding would severely stall U.S. cancer research, threaten future breakthroughs, and push young scientists out of the field.
🔹 In his Substack, Eric Topol reviews four recent studies linking immune aging and chronic inflammation to age-related diseases, presenting a synthesis in which declining immune resilience plays a central role in reduced healthspan.
🔹 The London Biotechnology Show returns June 18–19, 2025 at ExCeL London, expecting 3,500+ attendees and 1,800 biotech firms; highlights include AI-driven bioprocessing, precision medicine, and cell/gene therapies, with keynotes from leaders at IBM, Merck, Microsoft, MHRA, and Novo Nordisk.
🚀 A New Kid on the Block
(Emerging startups with a focus on technology)
🔹 Kiin Bio raises $2.2M pre-seed, led by b2venture, to develop AI-powered "virtual scientists" that coordinate drug discovery workflows to streamline pharma R&D without expanding headcount, with tools priced per agent and built for integration.
This newsletter reaches over 8.8K industry professionals from leading organizations across the globe. Interested in sponsoring?
Contact us at info@biopharmatrend.com
MIT & Recursion Open-Source New Joint Structure–Affinity Prediction Model
MIT’s Jameel Clinic and CSAIL, in collaboration with Recursion, have launched a biomolecular foundation model designed to predict both 3D protein structures and their binding affinities. Built on NVIDIA’s BioHive-2 infrastructure and extending Boltz-1 release, Boltz-2 reportedly matches free energy perturbation (FEP) methods in accuracy while running over 1000 times faster.

The model integrates ~5 million affinity measurements, molecular dynamics (MD) simulations, and structural distillations into a unified architecture. Notable updates include a dedicated affinity module, a Boltz-Steering method to enhance physical realism, and user-configurable constraints for guided modeling. Unlike approaches that treat structure and affinity as separate steps, Boltz-2 predicts both jointly—achieving top performance in CASP16 on 140 complexes, and generalizing across protein–nucleic acid and antibody–antigen systems.
MIT researchers Regina Barzilay and Tommi Jaakkola led development, with contributions from Gabriele Corso, Saro Passaro, Jeremy Wohlwend, and teams at MIT and Recursion. The team is presenting Boltz-2 at MIT (June 9), San Francisco and GTC25 Paris (June 11), and MoML Montreal (June 17).
The model is under the MIT license: boltz.bio/boltz2
We initially reviewed biological foundation models a year ago in “19 Companies Pioneering AI Foundation Models in Pharma and Biotech” and revisited this March with “13 Foundation Models: Startups, Industry Updates and the Nobel Prize”; Boltz-2 is now included in our recent June addition.
Spatial Biology Means Handling Terabyte-Scale Files
Kenny Workman, co-founder and CTO at LatchBio, recently outlined some of the raw data demands emerging from spatial biology workflows. Referencing a CosMX SMI experiment on a 100mm² section of liver cancer tissue, he listed approximate data volumes per modality:
~50GB for spatially-resolved metadata from 464,000 cells
~300GB for 534 million transcripts
<1GB for cell boundary polygons
~500GB for raw and stitched morphology images
Altogether, a single run can approach 1TB.

Workman says working with these datasets needs infrastructure that can quickly search, filter, and render complex image and molecular data, something traditional databases often can’t handle well.
A Visual Guide to Genome Editors
Evan DeTurk, a CRISPR researcher at UC Berkeley, has published a visual guide to genome editing tools on Asimov Press, aimed at readers seeking a foundational understanding of today’s rapidly expanding editing systems. The piece walks through the origins, mechanisms, and known limitations of several key genome editors: Cas9, Cas12, Cas13, base editors, prime editors, and the recently described bridge RNA system.
The guide also outlines off-target effects for each system and explains where certain editors (like Cas12 and base editors) may offer improvements in specificity or precision over Cas9. The inclusion of emerging systems like bridge RNA and TIGR-Tas suggests a broadening frontier, where genome editing is no longer synonymous with CRISPR alone.
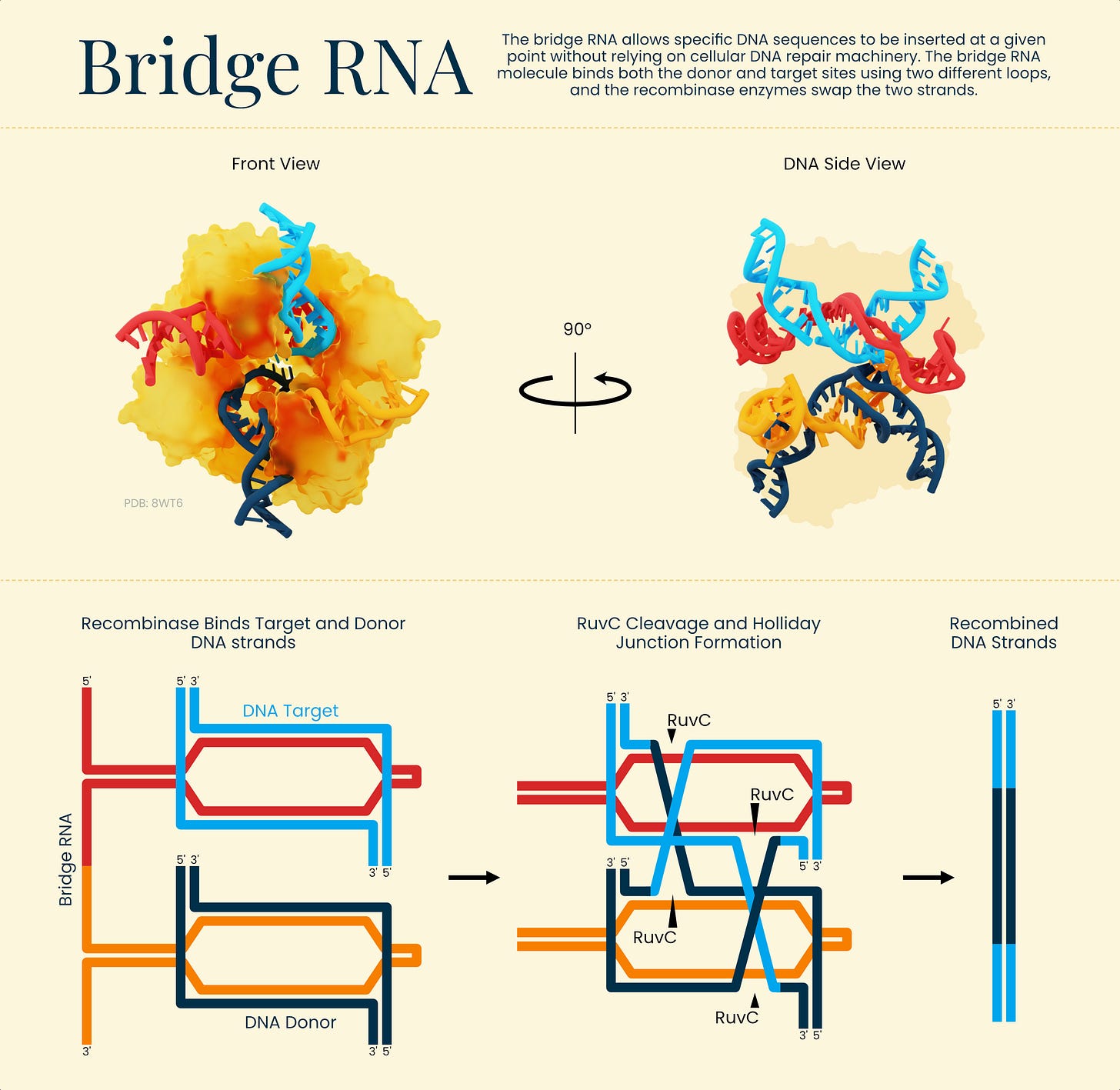
Deep Learning: Pros & Cons in Digital Pathology
A spring paper in Annals of Oncology takes a wide-angle look at how AI is being used in cancer care, especially in pathology. While deep learning tools can now identify tumors, classify cancer subtypes, and even predict molecular features directly from slide images, most of these systems are still far from routine clinical use. No AI-driven biomarkers, for instance, have yet cleared the bar for top-tier clinical evidence.
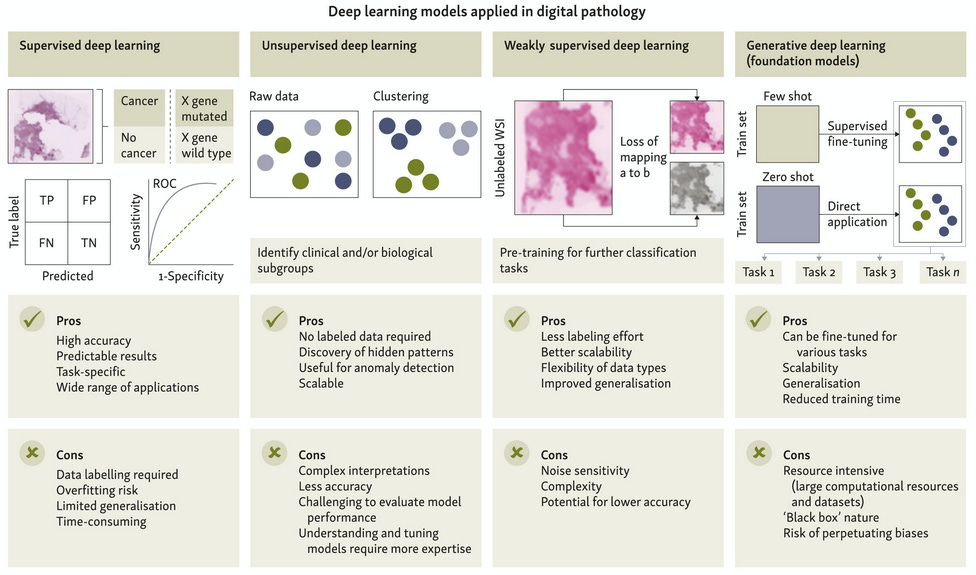
The paper also digs into why adoption has been slow. Some reasons are technical, like overfitting or inconsistent staining in slides, but others are human—clinicians wary of opaque “black box” models, regulatory frameworks still catching up, and the reality that many models work best on well-studied cancers with ample training data.
Still, there’s momentum around newer directions, like models that combine multiple data types (genomics, pathology, clinical notes) to give more useful predictions. The hope is that these generalist systems might eventually help fill in diagnostic gaps and support decisions, but that will take clearer standards and more real-world validation.
Biotech VC Enters Quieter Phase
On BioSpace, senior editor Annalee Armstrong recaps a new Pitchbook report marking the end of biotech’s 2012–2024 venture cycle—one marked by early growth, pandemic-fueled acceleration, and a sharp contraction. With fundraising levels now back near 2012 baselines, the sector appears to be entering a more restrained, long-term phase of company formation and capital deployment.

According to Annalee’s reading of the report, specialist firms like Flagship Pioneering, Atlas, and 5AM Ventures continue to build new companies but with tighter budgets and an emphasis on platform models. Generalists like Lux and Dimensions may benefit from lower valuations and less crowding. Exit activity remains limited, prompting funds to plan for longer holding periods. Still, capital-efficient startups like Maze Therapeutics and Artbio show there's room to advance even under leaner conditions.
Why AI Hallucinations May Be a Topological Problem
Derrick Hodge argues that AI hallucinations stem from topological flaws in transformer architecture rather than from the training data. Drawing on topological and algebraic concepts, Hodge suggests that transformer-based models function entirely in continuous spaces, where concepts blend too easily. This, he argues, makes it mathematically possible for models to generate confident but incorrect statements by smoothly interpolating between unrelated ideas.
Hodge proposes that adding torsion (a property from differential geometry) could enforce discrete separations between concept classes and prevent invalid inferences. His forthcoming paper, Semantic Rings and Concept Formation in Transformers, set for release June 25, will reportedly explore how semantic structure, including loops and sheaf cohomology, might be used to formalize and constrain meaning in AI systems.
✨ Research highlight: Apple study finds that long-context reasoning models fail abruptly on complex structured tasks—collapsing into incoherent outputs and often underperforming smaller LLMs on simpler problems (though some argue these failures stem from task setup and context limits, not reasoning flaws), raising questions about their claimed reasoning generalization.
Read also:
13 Foundation Models: Startups, Industry Updates and the Nobel Prize